In Vitro Kinase-Substrate ID Custom Kinase-Substrate Profiling (CKSP) Service
The vast majority of the proteins phosphorylated by specific protein kinases in humans and other species remain unknown despite more than 40 years of intense effort. We have identified at least 10,000 kinase-substrate phosphosite pairs from our literature searches, but we believe that actual number is closer to 10 million. With the emergence of protein kinases as one of the most promising families of drug targets in the pharmaceutical industry today, it is critical to define the proteins that are controlled by these important regulatory enzymes. Our Custom Kinase Substrate Profiling (CKSP) Service uniquely permits the rapid simultaneous identification of a panel of physiological substrates for most known protein kinases in any tissues or cells of specific interest. Our cost effective strategy also provides for the identification of the specific sites at which these substrates are phosphorylated, as well as commercial antibodies that can be used to specifically track and quantify these phosphosites in follow up studies. Our methodology is superior to any other procedures that have been applied to kinase substrate identification including mass spectrometry and protein microarrays, which are more expensive and do not define reagents for follow on experiments.
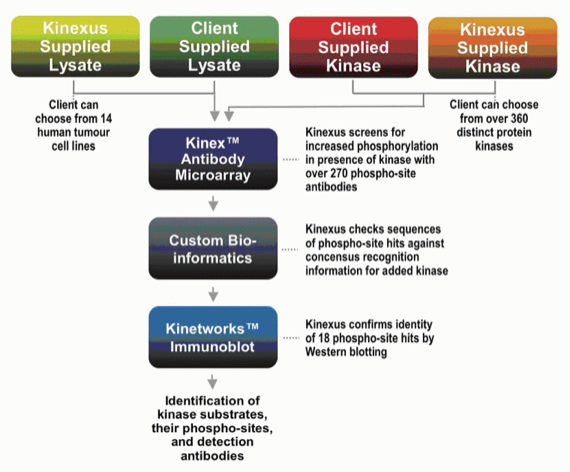
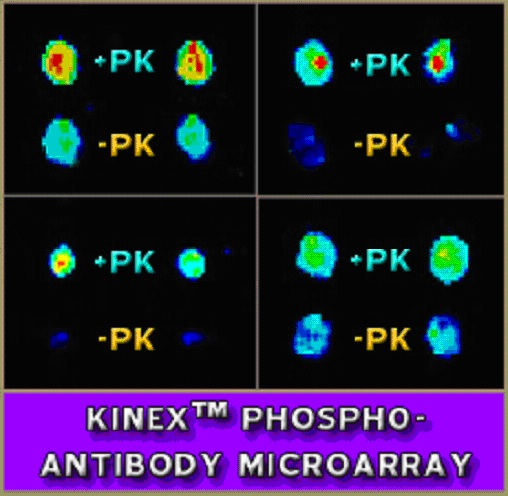
With our flexible and convenient protein kinase substrate identification service, clients can send us their own kinases and cell/tissue lysates or they can choose from our collections of over 360 active human kinases, and lysates from 14 of the most commonly studied human tumour cell lines. Within four weeks of receipt of a client order, Kinexus will provide information about a large panel of potential substrates as revealed with a ~350 phosphosite antibody microarray and validation studies with 18 of the most promising leads by immunoblotting. The selection of the candidate phosphosites for the Western blotting exploits proprietary information that Kinexus has developed to determine negative and positive determinants for specific kinase substrate recognition. The results from this unique service can be used to define signalling pathways impacted by specific kinases, identify reagents to enable kinase inhibitor discovery in vitro, and ascertain the effectiveness and specificity of kinase inhibitor drugs in living systems.
We have compiled data on more than 960,000 known and putative phosphosites in over 21,000 human proteins, and this is available in our PhosphoNET KnowledgeBase. With at least 536 known human protein kinases, each kinase appears to target over 1000 phosphosites on average. At this juncture, for over a third of the human protein kinases, not a single physiological substrate has been reported in the scientific literature.
With our flexible and convenient protein kinase substrate identification service, clients can send us their own kinases and cell/tissue lysates or they can choose from our collections of over 360 active human kinases, and lysates from 14 of the most commonly studied human tumour cell lines. Within four weeks of receipt of a client order, Kinexus will provide information about a large panel of potential substrates as revealed with a ~350 phosphosite antibody microarray and validation studies with 18 of the most promising leads by immunoblotting. The selection of the candidate phosphosites for the Western blotting exploits proprietary information that Kinexus has developed to determine negative and positive determinants for specific kinase substrate recognition. The results from this unique service can be used to define signalling pathways impacted by specific kinases, identify reagents to enable kinase inhibitor discovery in vitro, and ascertain the effectiveness and specificity of kinase inhibitor drugs in living systems.
We have compiled data on more than 960,000 known and putative phosphosites in over 21,000 human proteins, and this is available in our PhosphoNET KnowledgeBase. With at least 536 known human protein kinases, each kinase appears to target over 1000 phosphosites on average. At this juncture, for over a third of the human protein kinases, not a single physiological substrate has been reported in the scientific literature.
Furthermore, for the majority of the remaining human protein kinases, only one or two phosphoprotein substrates have been identified. A comprehensive understanding of the composition and architecture of cell signalling networks will require a much deeper knowledge of which phosphoproteins and more specifically which of their phosphosites are targeted by certain protein kinases, and what is the physiological consequence of their phosphorylation.
Most of the known phosphorylation sites have been elucidated through tandem MS-MS mass spectrometry. This powerful and sensitive method relies on phosphoprotein enrichment techniques such as strong cation exchange chromatography and can permit identification of hundreds of phosphosites at once. However, it is not very quantitative, and it is impractical and costly for routine analysis of phosphorylation changes in tissues and cells that are often limiting in quantity. Moreover, with a novel phosphorylation site defined by mass spectrometry it can take more than 6 months at high expense to develop a reliable phosphosite-specific antibody for its detection by immunoblotting and immunohistochemistry. With the availability of several hundred commercial antibodies from several vendors, Kinexus has reversed the paradigm for phosphosite discovery with its Kinex™ antibody microarray and Kinetworks™ multi-immunoblotting services. With these methodologies, Kinexus uses panels of phosphosite antibodies to detect known and cross-reactive proteins that display altered phosphorylation in living systems in response to hormonal and pharmacological manipulations or, as with our Custom Kinase Substrate Profiling Service, increased phosphorylation in vitro with purified kinases.
Any cross-reactive proteins identified with our combined microarray and immunoblotting services can be easily enriched with the detecting phosphosite antibody and identified by Tandem MS/MS mass spectrometry. Kinexus is pleased to offer this protein fingerprinting technique to our clients with our Protein Identification by Mass Spectrometry (PIMS) Services. Moreover, since the phosphosite epitope of the detection antibody is known, it is often easy to locate the affected phosphorylation site without further sequencing. Thus antibody-driven phosphoprotein discovery is highly cost effective, informative and enabling for immediate follow up analyses with other methodologies that exploit antibodies. With our Custom Kinase Substrate Profiling Service, for their favourite experimental model system and a kinase supplied by Kinexus, any researcher can have a panel of physiological substrates, the phosphosites, and detecting phosphosite antibodies identified for less than US$3000.
Most of the known phosphorylation sites have been elucidated through tandem MS-MS mass spectrometry. This powerful and sensitive method relies on phosphoprotein enrichment techniques such as strong cation exchange chromatography and can permit identification of hundreds of phosphosites at once. However, it is not very quantitative, and it is impractical and costly for routine analysis of phosphorylation changes in tissues and cells that are often limiting in quantity. Moreover, with a novel phosphorylation site defined by mass spectrometry it can take more than 6 months at high expense to develop a reliable phosphosite-specific antibody for its detection by immunoblotting and immunohistochemistry. With the availability of several hundred commercial antibodies from several vendors, Kinexus has reversed the paradigm for phosphosite discovery with its Kinex™ antibody microarray and Kinetworks™ multi-immunoblotting services. With these methodologies, Kinexus uses panels of phosphosite antibodies to detect known and cross-reactive proteins that display altered phosphorylation in living systems in response to hormonal and pharmacological manipulations or, as with our Custom Kinase Substrate Profiling Service, increased phosphorylation in vitro with purified kinases.
Any cross-reactive proteins identified with our combined microarray and immunoblotting services can be easily enriched with the detecting phosphosite antibody and identified by Tandem MS/MS mass spectrometry. Kinexus is pleased to offer this protein fingerprinting technique to our clients with our Protein Identification by Mass Spectrometry (PIMS) Services. Moreover, since the phosphosite epitope of the detection antibody is known, it is often easy to locate the affected phosphorylation site without further sequencing. Thus antibody-driven phosphoprotein discovery is highly cost effective, informative and enabling for immediate follow up analyses with other methodologies that exploit antibodies. With our Custom Kinase Substrate Profiling Service, for their favourite experimental model system and a kinase supplied by Kinexus, any researcher can have a panel of physiological substrates, the phosphosites, and detecting phosphosite antibodies identified for less than US$3000.